The SciKnowMine Triage Application
We present here a user manual for running and maintaining a web-based system for peforming document triage given a corpus of PDF files. We will describe processes for installation, execution and maintenance of the system.
- Installation Manual
- System Organization
- Command Line - Set up
- Command Line - Working with Data
- Command Line - Reporting Functions
- Command Line - Deleting Data
- Command Line - Machine Learning
- Command Line Tools - Running Experiments
- Web Application - Running the System
- Web Application - Extracting text using LAPDF-Text
- Web Application - Performing the triage task
- Web Application - The Base Digital Library
12. Web Application - The Base Digital Library
Once the documents have been selected for inclusion in a particular corpus, then more detailed information extraction and annotation can be performed. This work is still at an early stage but is intended as a core functionality for the SciKnowMine system. In particular, we anticipate using Information Extraction to locate and extract passages of interest within the system.
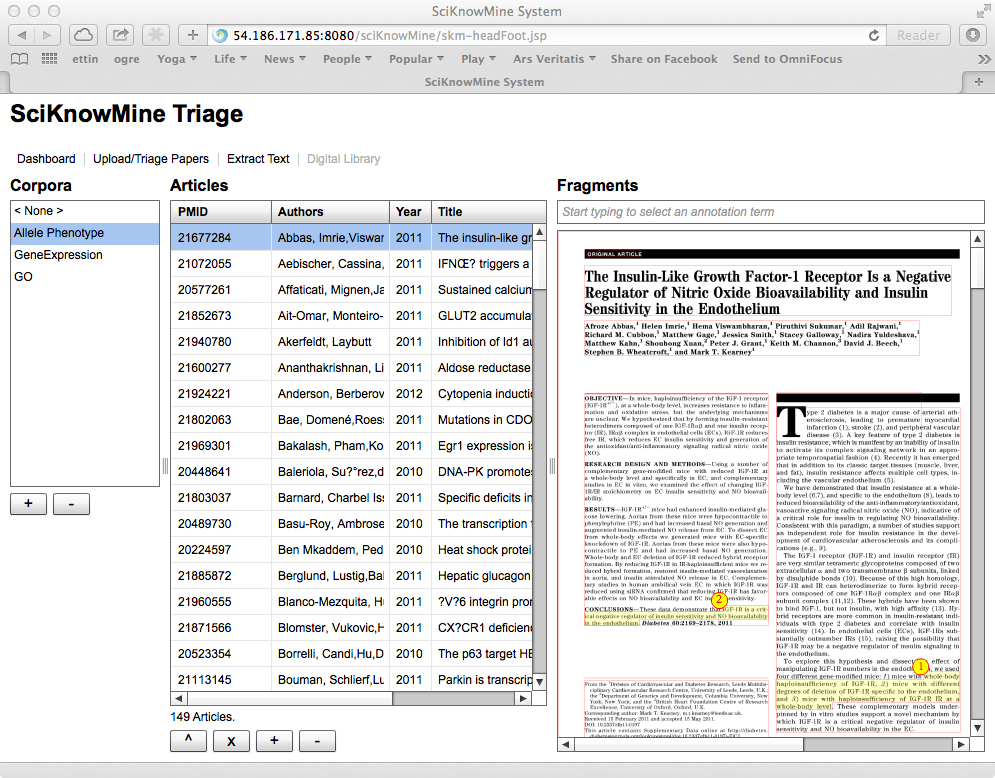
The screenshot shows a specific target corpus selected for the MGI-driven use-case:
'Allele Phenotype'. The user may annotate passages in the text by clicking, dragging
and releasing the mouse over the document. These two selected passages shown are the
main claims of the paper shown. In particular, we anticipate the use of the BioC
annotation standard to support sharing and structuring
these fragment elements.
By focusing on PDF files as a common de-facto publication standard that most scientists are comfortable using, we hope to develop knowledge engineering and text mining applications that scientists find intuitively useful in the execution of their day-to-day work. We also intend that our tools are portable and modular so that they may be incorporated into other developers' systems.