Developing NL Annotations for KEfED Elements
Here we describe the first practical implementation of a system to develop KEfED models directly from the text of a research article.
June 2016
July 2015
May 2015
April 2015
January 2015
- Coprecipitation Frames v2
- List of Experimental Motif Types + Definitions
- KEfED Database Construction
December 2014
October 2014
- Pathway Logic Experiment Types
- Building a Database of Observations from Result Text
- Deploying the BioScholar System
- Reading Against a Model of Experimental Evidence
August 2014
-
Fragments and KEfED models
We drive the definition of simple KEfED models through the Fragmenter. We denote each fragment by a code (e.g., ‘Fig5C’’), that here refers to the figure but could conceivably be based on any label for the model.
Here, we show a screenshot of the system in its current form with a simple KEfED model based on the experimental protocol used to generate the data of Figure 5C from Montero Conde et al. 2013. The system shows the text of the fragment on-screen and updates the underlying database entry as you enter individual elements and edges into the system. Importantly, the Data Structure panel on the right hand side shows the variable dependencies for the currently selected element (in other words, there each individual measurement of Relative Light Units derived from the this more ). We will incorporate this piece to model more complex data structures derived from statistical analysis and data processing steps.
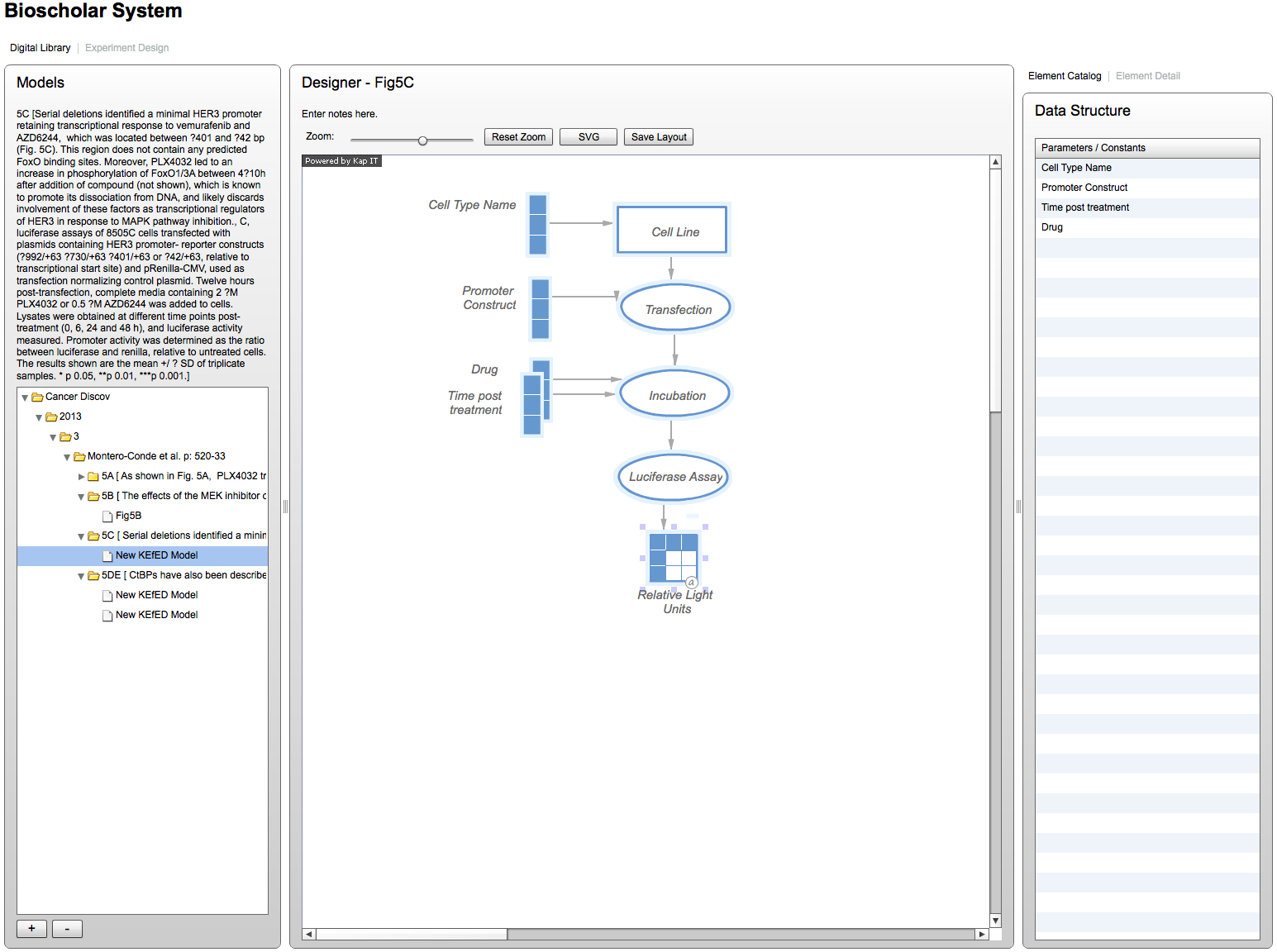 Figure 1: Screenshot of KEfED model
Figure 1: Screenshot of KEfED model 5c from Montero Conde et al. 2013.
Note that the actual text of the Figure legend reads:
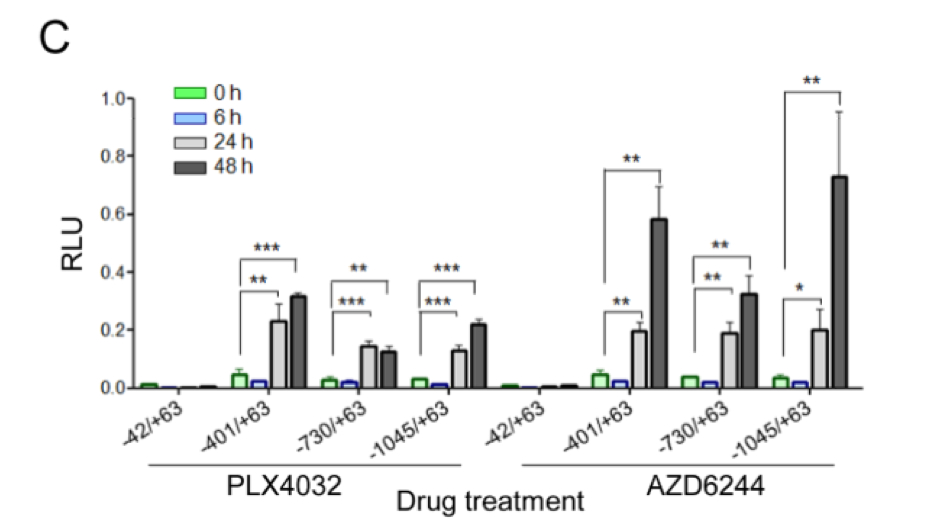
C, luciferase assays of 8505C cells transfected with plasmids containing HER3 promoter- reporter constructs (−992/+63; −730/+63; −401/+63 or −42/+63, relative to transcriptional start site) and pRenilla-CMV, used as transfection normalizing control plasmid. Twelve hours post-transfection, complete media containing 2 μM PLX4032 or 0.5 μM AZD6244 was added to cells. Lysates were obtained at different time points post-treatment (0, 6, 24 and 48 h), and luciferase activity measured. Promoter activity was determined as the ratio between luciferase and renilla, relative to untreated cells. The results shown are the mean +/ − SD of triplicate samples. * p< 0.05, *p < 0.01, **p < 0.001
This illustrates that the text we capture from the PDF file is somewhat inaccurate (but is the only text source we have for this paper since it is not part of the PubMedCentral OA set, but we could conceivably scrape the online HTML on a paper-by-paper basis, requiring getting permission from the publishers).
… and the figure portraying the data itself in the paper is this:
It is immediately evident from the figure that the measurements are based on Relative Light Units (RLU), and the axes of experiment are (a) the Cell Type Name, (b) Promoter Construct, (c) Time after treatment and (d) the Drug.
Our long term challenge is (A) to be able to infer or select this design based only on the data presented in the scientific paper, (B) populate this model with values derived from the content of the paper and then (C) to interpret this data correctly to support reasoning about cancer pathways. Note that this sort of reasoning need not conform to ‘traditional’ modeling approaches but may leverage Bayesian / Causal reasoning based on the the variables that are directly implicated in the experimental design or hidden ‘interpretive’ variables of importance to the domain.
We have gone through each of the experiments in Figure 5 of Montero-Conde et al. 2013 from parts A-F, decomposing the text in detail to infer the underlying data and methods used to substantiate the claims made in the paper. This forms the basis of our continued discussion with the Pathway Logic Datum formalism (available here: http://light.csl.sri.com/datum) and possible connections to other formalisms such as BEL (http://www.openbel.org/).
Montero-Conde-2013-3-520-figure5A-F.pdf
We simply go through five assertions from the narrative text of the document describing some findings of the paper reported in a single figure. In fact, these statements are derived from six separate small scale experiments which together form the basis of the evidence pertaining to this section of the paper.
-
KEfED-based NL Annotations
The current annotation scheme for extracting relations from text into the KEfED model is shown below:
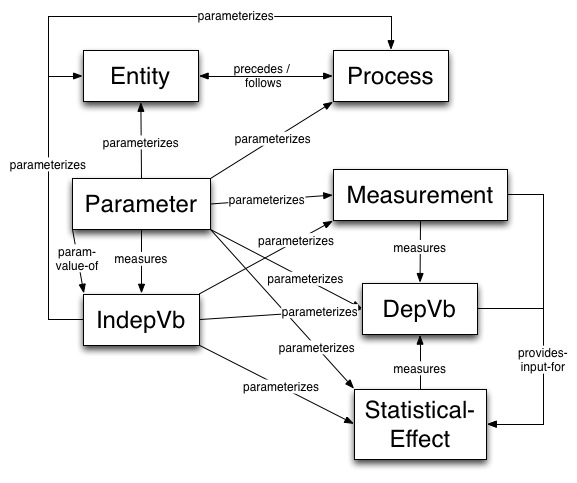
This currently shows the schema used in brat to annotate sentences for NLP. Here, we focus mainly on the relationships between the Statistical Effect, Dependent Variable and Parameter classes as a preliminary structure to be able to extract from text the dependency relations that lie at the heart of the KEfED representation of the structure of data cited in systems biology experiments.
We are currently in the process of curating many such sentences in order to then track how these annotations (A) reflect readable linguistic features from within the text and (B) correctly reconstruct the parameterization signature of experimental findings based on the KEfED model. Formulating and evaluating this structure is a key contribution of this effort.