The SciKnowMine Triage Application
We present here a user manual for running and maintaining a web-based system for peforming document triage given a corpus of PDF files. We will describe processes for installation, execution and maintenance of the system.
- Installation Manual
- System Organization
- Command Line - Set up
- Command Line - Working with Data
- Command Line - Reporting Functions
- Command Line - Deleting Data
- Command Line - Machine Learning
- Command Line Tools - Running Experiments
- Web Application - Running the System
- Web Application - Extracting text using LAPDF-Text
- Web Application - Performing the triage task
- Web Application - The Base Digital Library
11. Web Application - Performing the triage task
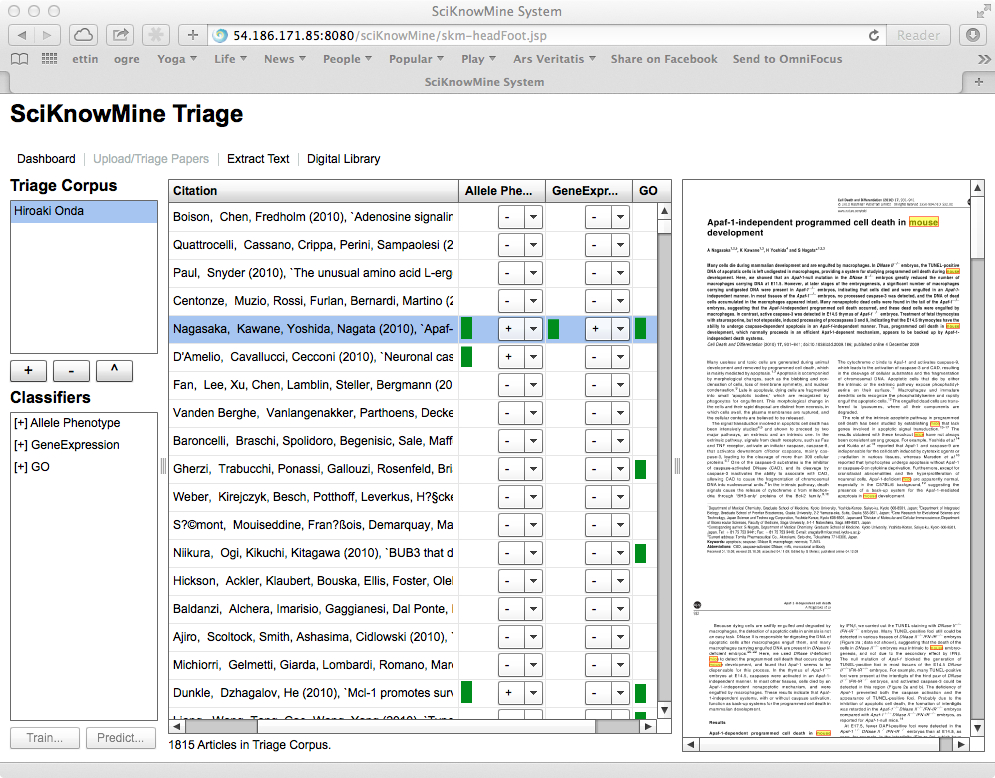
The goal of this application is to accelerate the process of document triage for biocurators by providing document classification cues to the biocurator using the system. Within the tool, this consists of listing the documents so that they can be scored easily and showing cues to features within each document that support the classification.
The triage component requires that the user select a Triage Corpus (such as Hiroaki
Onda as shown), which then lists the documents in the corpus, cross-referenced to inclusions
codes for each corpus in the system (+, - or ?).
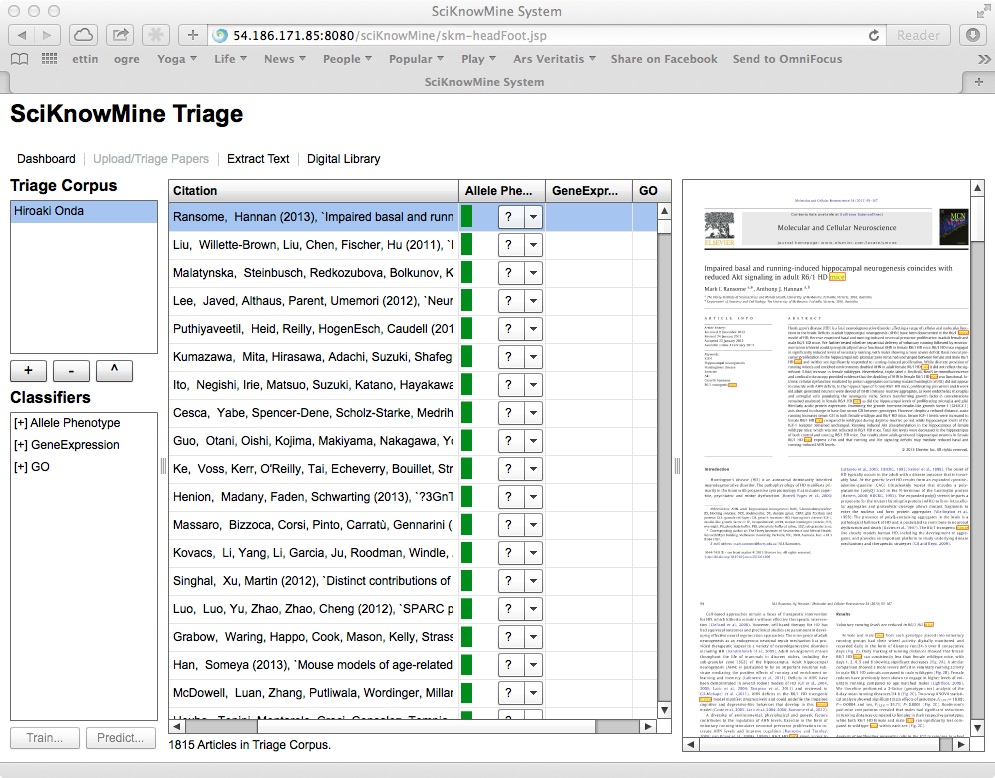
If a trained classifier has been run on the documents in a given triage corpus, a score will have been generated for the document in this context. This shows as a green bar next to the code selection. The system also permits the user to sort the columns of the table in descending order of this score by clicking on a triage corpus heading (and the system removes all other data for the documents shown). This is shown below.
The selected paper shown is a previously-unseen document with the highest classification
score available. The view of the PDF file shows the body of the PDF file with all
instances of the words mouse, mice or murine highlighted. This acts as an important
cue for Mouse Genome Informatics curators and will be changed when the system is applied
to other datasets.
Training and running the classifiers from within the system at the present time is
based only on an SVMLite bigram classifier. In order to train a classifier, the user must
select a Corpus name (the target of the classification) and click the Train... button.
The system will create a modal feedback popup and report on execution progress since this
step takes time. Similarly, to use the classifier to predict inclusion of individual
papers, the user must select a both the Corpus's classifier and a Triage Corpus to run
over before clicking the Predict... button.